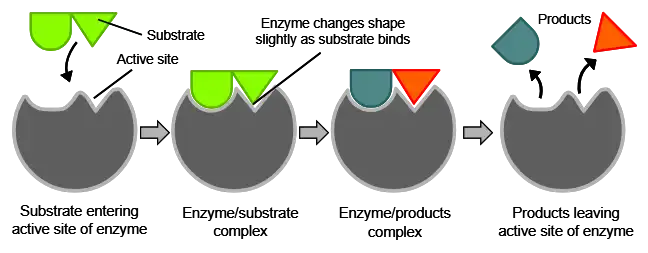
Enzymes are essential biological catalysts that speed up chemical reactions in living organisms without being altered themselves. This article provides a thorough overview of enzyme activity, including the critical roles of enzymes, enzyme classification systems, kinetics, common assays, and the various factors influencing enzyme function.
What are Enzymes?
Enzymes are primarily proteins that catalyze or increase the rate of biochemical reactions. The enzyme itself remains unchanged by the reaction and can be reused multiple times.
Without enzymes, many essential chemical transformations in cells would proceed extremely slowly or not at all.
Enzymes have an active site that provides a unique chemical environment to bind to specific substrates. This active site perfectly complements the substrate molecular structure and allows enzymatic catalysis to take place.
The substrate(s) binds to the active site through multiple weak interactions like hydrogen bonding, resulting in the enzyme-substrate complex.
In addition to proteins, a small subset of enzymes comprises catalytic RNA molecules called ribozymes. Examples include ribonuclease P and the hammerhead ribozyme. However, protein-based enzymes are far more abundant and diverse.
The Roles of Enzymes in Cells and Organisms
Enzymes carry out a vast array of essential functions in sustaining life. Some key examples include:
- Metabolic reactions: Enzymes catalyze all the complex sequences of metabolic pathways like glycolysis, Krebs cycle, electron transport chain, etc., that generate energy and synthesize important biomolecules.
- DNA replication: Helicases unwind the DNA double helix, while polymerases catalyze the synthesis of new complementary strands during replication.
- Gene expression: RNA polymerases synthesize RNA transcripts from DNA genes. Other enzymes process and splice RNA.
- Cell signaling: Kinases and phosphatases add or remove phosphate groups to regulate signaling proteins.
- Cell motility: Motor proteins like kinesin and dynein generate mechanical force through ATP hydrolysis to transport cargo within cells.
- Apoptosis: Caspases cleave regulatory and structural proteins as part of the programmed cell death pathway.
- Blood clotting: Thrombin, fibrin, and other enzymes trigger a cascade response to form blood clots upon injury.
- Digestion: Digestive enzymes like amylases, proteases, and lipases break down food polymers into absorbable molecules.
As evident, enzymes control practically every biological process and allow organisms to grow, reproduce, and sustain homeostasis. Defects in enzymes lead to metabolic disorders and diseases.
Enzyme Classification Systems
Enzymes are divided into different classes based on the type of reactions they catalyze. The International Union of Biochemistry (IUB) has developed a classification system that assigns each enzyme a 4-digit EC number.
The EC number indicates the enzyme class, subclass, sub-subclass, and serial number. For instance, lysozyme, which cleaves glycosidic bonds in bacterial cell walls, is designated as EC 3.2.1.17, where:
- 3 indicates it is a hydrolase (catalyzes hydrolysis reactions)
- 2 indicates it acts on glycosyl compounds
- 1 indicates it is a glycosidase
- 17 is the serial number of the specific enzyme in the sub-subclass
The 6 main IUB enzyme classes are:
- Oxidoreductases: Catalyze oxidation-reduction reactions involving electron or hydrogen transfer between molecules. Examples are dehydrogenases, peroxidases, and oxygenases.
- Transferases: Transfer functional groups like methyl, amino, and phosphate groups between substrates. Examples are kinases, transaminases, transmethylases.
- Hydrolases: Catalyze hydrolysis reactions that cleave bonds using water. Examples are proteases, lipases, phosphatases.
- Lyases: Catalyze non-hydrolytic addition or removal of groups from substrates. Examples are decarboxylases and dehydratases.
- Isomerases: Catalyze structural rearrangements within a single molecule. Examples are racemases and epimerases.
- Ligases: Catalyze bond formation between substrates by condensation reactions coupled to ATP hydrolysis. Examples are DNA ligase and acetyl CoA synthetase.
Alternatively, enzymes can also be classified based on the type of substrate they act upon like:
- Proteases act on protein substrates
- Lipases act on lipid substrates
- Polymerases act on DNA/RNA polymers
- Kinases act on proteins by phosphorylating them
Enzyme Kinetics
Enzyme kinetics refers to measuring reaction rates and analyzing how kinetic parameters change with varying substrate/enzyme levels. This provides valuable insights into the catalytic mechanism and efficiency.
Key kinetic parameters are:
- Maximum velocity (Vmax): Maximum rate of the enzymatic reaction when the enzyme is saturated with substrate.
- Michaelis-Menten constant (Km): Substrate concentration at which the reaction rate is half Vmax. Indicates substrate-binding affinity.
- Turnover number (kcat): Maximum number of substrate molecules converted to product per enzyme active site per second. Indicates catalytic efficiency.
Enzyme kinetics is studied by measuring initial reaction rates at different substrate concentrations. A Michaelis-Menten kinetic plot depicts the hyperbolic relationship between reaction rate and substrate concentration.
Km and Vmax values are derived from such plots. Lower Km indicates higher substrate affinity, while higher Kcat indicates superior turnover capacity. High kcat/Km ratios signal enzymes with great catalytic power.
Common Enzyme Activity Assays
There are several assays to measure enzyme activity by monitoring reaction progress. Common methods include:
- Spectrophotometry: Measures absorbance changes as substrates are converted to products. Works for colored substrates/products.
- Fluorometry: Monitors fluorescence changes during reactions. Fluorescent substrates/products are used.
- Chromatography: Reaction components are physically separated and quantified. Works for non-colored substrates.
- Radiometry: Radiolabeled substrates are tracked throughout reactions. Sensitive, quantitative detection.
- Electrochemistry: Electron transfer during redox reactions is monitored using electrodes.
- Turbidometry: Measures changes in solution turbidity as insoluble products form.
- pH change: Hydrogen ion concentration changes can indicate reaction progress.
Assays should be designed to ensure initial rate conditions using low enzyme levels and excess substrate. Activity is calculated from the initial linear increase in product formation over time.
High-throughput screening uses automated assays to rapidly test enzyme variants or drug candidates that affect specific enzymes.
Factors Influencing Enzyme Activity
Many chemical and physical factors in the cellular milieu can affect enzyme activity and kinetics:
1. Temperature
- Enzyme reaction rates increase with rising temperatures as kinetic energy and molecular collisions increase until an optimum temperature is reached.
- Each enzyme has an ideal temperature for maximum catalytic activity, generally between 35-45°C for human enzymes.
- At lower temperatures, enzyme activity is reduced due to decreased molecular motions and flexibility of the enzyme structure.
- However, above the optimum temperature, further heating begins to denature the 3D structure of the enzyme protein due to thermal instability.
- High temperatures break the weak bonds like hydrogen bonds and hydrophobic interactions that maintain folded conformation. This leads to unraveling and loss of active site geometry.
- Enzymes from thermophilic organisms have higher optimum temperatures due to specialized stabilizing structural features. But most enzymes become inactive by 70-80°C.
- Temperature not only affects the rate of enzymatic reactions but also the equilibrium position. The equilibrium constant of exothermic reactions decreases with rise in temperature.
2. pH
- Each enzyme shows maximal catalytic activity at a specific pH known as the optimum pH. This is typically between pH 5-9 for most enzymes.
- Extremes of very high or low pH values usually lead to loss of proper enzyme structure and function due to denaturation.
- pH influences the ionization states of key amino acids involved in substrate binding and catalysis.
- Acidic or basic pH can protonate or deprotonate groups essential for activity. Electrostatic interactions are affected.
- Active site geometry depends on ion pairs formed between positively and negatively charged amino acids. Changing pH disrupts these critical ion pairs.
- Some enzymes, like pepsin, have evolved to function in highly acidic conditions, like a stomach pH of 1-3.
- Likewise, detergent enzymes work well in an alkaline pH of 8-10. But such extremes denature most other enzymes.
- The pH optimum varies based on where the enzyme normally functions. Cytoplasmic enzymes have optima near neutral pH, while digestive enzymes are optimized for acidic secretions.
- Buffers are used to maintain the pH close to the enzyme optimum both in vivo and in vitro experimental conditions.
3. Substrate Concentration
- The reaction rate increases proportionally as the substrate concentration rises, providing more substrate molecules to bind to the active sites.
- However, at very high substrate concentrations, the enzyme active sites eventually get saturated as all sites are occupied.
- Beyond saturation, a further increase in substrate levels does not increase the rate since no new enzyme-substrate complexes can form.
- The maximum reaction rate or Vmax is achieved at substrate concentrations far above the enzyme’s Km value, indicating complete saturation.
- The substrate concentration for reaching Vmax depends on the enzyme-substrate binding affinity. Stronger binding enzymes require a lower substrate for saturation.
- Weak inhibitor compounds can also act as substrates at very high concentrations and elicit Vmax rates by overwhelming the inhibition.
- Substrate inhibition may occur at excessively high substrate concentrations due to multiple substrate molecules jamming active sites.
4. Enzyme Concentration
- The reaction rate is directly proportional to the enzyme concentration when other conditions, like substrate levels, are non-limiting.
- Having more enzyme molecules in the assay increases the total number of active sites available.
- Each active site can convert substrate to product at its kcat rate. So, higher enzyme levels linearly increase the overall reaction rate.
- This relationship helps determine enzyme levels needed for desired catalytic capacity based on substrate amounts.
- In cells, the total enzyme concentration is regulated at adequate levels to support flux through metabolic pathways.
- Enzyme production rates are controlled by gene expression regulation, translation, and protein degradation.
5. Inhibitors
- Inhibitors are compounds that bind to enzymes and reduce their catalytic activity.
- Competitive inhibitors occupy the enzyme’s active site, competing with the substrate for binding. This obstructs substrate access to the site.
- Increasing substrate concentration can overcome this inhibition by out-competing the inhibitor.
- Non-competitive inhibitors bind at allosteric sites away from the active site. This changes enzyme shape and distorts the active site, reducing function.
- Uncompetitive inhibitors only bind to the enzyme-substrate complex, not the free enzyme. This also lowers activity.
- Irreversible or covalent inhibitors form permanent chemical bonds to active site residues. This irreversibly inactivates the enzyme.
- Most inhibitors display mixed inhibition, combining aspects of multiple modes. Inhibitors are used clinically to target key pathogens or disease enzymes.
6. Activators
- Enzyme activators are compounds that increase the catalytic activity of enzymes.
- Activators typically bind to specific allosteric sites on the enzyme, distinct from the active site.
- This induces a conformational change in enzyme shape that enhances its affinity for the substrate at the active site.
- The activator may also facilitate product release, allowing more rapid enzyme turnover.
- This modulation of activity provides a means of regulating metabolic flux through pathways in response to cellular needs.
- Enzyme cofactors like metal ions can act as activators by assisting in substrate binding and transition state stabilization.
- Oxidative enzymes generally require activating metal ions. The activator may also be a protein subunit in a multi-enzyme complex.
Models Explaining Enzyme Structure and Catalysis
Over the years, different models have been proposed to explain enzyme structure, substrate binding, and catalytic mechanism:
Lock and Key Model
- First proposed by Emil Fischer in the 1890s based on shape complementarity.
- Envisioned the enzyme active site as having a rigid, predefined shape that fits only specific substrate molecules, akin to a lock and key.
- The binding of the substrate ‘key’ into the enzyme ‘lock’ occurs without any conformational changes.
- The lock and key analogy implies specificity but no induced fit. Only the correctly shaped substrate can bind to the active site.
- This model explains enzyme specificity well but does not account for kinetics or catalytic mechanisms beyond binding.
- Provides a simplistic preliminary explanation for selective substrate binding affinity seen in enzymes.
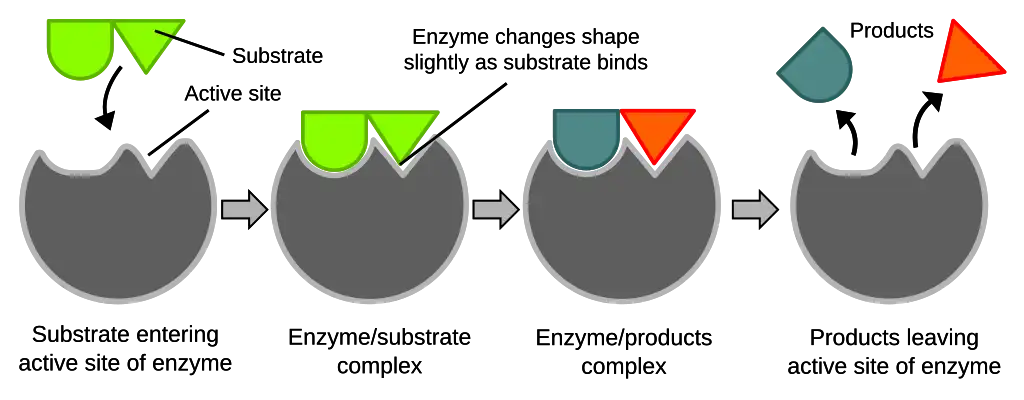
Induced Fit Model
- Proposed by Daniel Koshland in 1958 to modify some limitations of the lock and key model.
- The active site is envisioned as being flexible and undergoing conformational changes upon substrate binding.
- The substrate interaction causes the active site to mold around it and induce fit for optimal binding.
- This adjusts the active site shape and charges to orient and stabilize the transition state for catalysis properly.
- The induced fit enhances substrate affinity and allows catalytic groups to interact with the bound substrate.
- Accounts for conformational dynamics and substrate-induced changes contributing to enzyme function.
Cleft Model
- Suggests that the active site is a cleft or crevice rather than a pocket with defined boundaries.
- Substrate binding occurs through multiple weak interactions along the length of the cleft, not just within a pocket.
- It allows for some flexibility and movement, but the substrate itself does not change shape upon binding.
- Emphasizes distributed binding determinants rather than specific geometric complementarity seen in the lock and key model.
- An intermediate between very rigid and completely flexible interpretations of substrate binding.
Hand-Glove Model
- Conceptualized by Walter Kauzmann in 1959.
- An analogy of a hand fitting snugly into a glove; active site adjusts analogous to a glove around the substrate, which fits like a hand inserted into it.
- Combines aspects of both the rigid lock and key and flexible induced fit models.
- Some defined specificity exists, but the active site molds to the substrate like a glove.
- Accommodates subtleties of selective binding and optimized fit for transition state stabilization.
- No single model explains everything about enzyme kinetics and catalysis. The mechanism likely involves aspects of multiple models.
References and Sources
- Lehninger Principles of Biochemistry, 4th edition (David L. Nelson Michael M. Cox)
- 4.6 Enzymes – Human Biology. https://open.lib.umn.edu/humanbiology/chapter/4-6-enzymes/
- http://getfreeessays.com/effect-of-ph-and-temperature-enzyme-catalase-reaction-rate/
- Habte, Mezgebu Legesse, and Etsegenet Assefa Beyene. “Biological Application and Disease of Oxidoreductase Enzymes.” 2020, https://doi.org/10.5772/intechopen.93328.
- Green, Laura Kay. “In Vitro and in Vivo Characterisation of P. Aeruginosa Oxidoreductase Enzymes in Pathogenesis and Therapy.” 2012.
- Enzyme Inhibition. https://www.brainkart.com/article/Enzyme-Inhibition_27495/
- https://core.ac.uk/download/477734669.pdf.